Search Docs by Keyword
Getting Started with FASRC Storage
Getting Started with FASRC Storage
FASRC offers two complimentary storage offerings for groups:
- Lab Directory: holylabs
- path
/n/holylabs - 4TB (cannot be expanded)
- Retention allowed
- no backup (snapshots)
- path
- netscratch
- path
/n/netscratch - 50TB (cannot be expanded)
- 90-day retention policy
- no backup (snapshots)
- path
For more storage offerings, see the Data Storage Workflow documentation.
FASRC offers paid storage options, including Compute Storage, Lab Storage, and Long-Term storage (see the Data Storage Workflow documentation for more details). Your group may have purchased additional storage, so you could have other folders besides those on holylabs and netscratch.
How do I find my group’s storage folders?
Option 1: Ask a colleague
Typically, the easiest way to find out about your group’s storage is to ask a colleague.
Option 2: Use your group’s documentation
Many groups with FASRC maintain their own documentation. If your lab or group has internal documentation, it may indicate what storage you have access to.
Option 3: Self-service
The good news is you can also find the storage on your own!
- Familiarize yourself with one of our data management tools, Starfish, by reading the first three sections (Overview, Login, and Navigation) of the Starfish Zones Data Visualization Tool documentation.
- Connect to the FASRC VPN (Harvard VPN or a wired on-campus connection also work). For instructions on how to connect to the FASRC VPN, see the VPN Setup documentation.
- Go to the Starfish dashboard from any browser (Chrome and Firefox are preferred).
- To log in, use your FASRC username and FASRC password.
- Navigate to the group folder
- Right-click on the storage you would like to use
- Click on “Copy mount path to clipboard”
- Now, you have the storage “Path” in your clipboard. To keep it, paste it in a text editor, note-taker, or word processor.
/n/vast-holylabs/C/jharvard_lab - Very important! Note that the path above contains the letter
Cbeforejharvard_lab. The letterCindicates the filesystem is on Cannon cluster. If your group has a group share on FASSE, then you would seeF(instead ofC) before your group (<PI_lab>) to indicate it’s on the FASSE cluster. You must make two edits: removevast-and/C. The final path is:
/n/holylabs/jharvard_lab
How do I access my group storage?
Below, we describe various ways to access your group’s storage folders. You can use one or more methods that work well for you.
Option 1: Command-line
You can ssh to the cluster (see Terminal Access), then use the command cd (change directory) to your group share and the command ls (list) the contents of the folder
$ ssh jharvard@login.rc.fas.harvard.edu (jharvard@login.rc.fas.harvard.edu) Password: (jharvard@login.rc.fas.harvard.edu) VerificationCode: [jharvard@boslogin08 ~]$ cd /n/holylabs/jharvard_lab/ [jharvard@boslogin08 jharvard_lab]$ ls Everyone Lab [jharvard@boslogin08 jharvard_lab]$ cd Lab/ [jharvard@boslogin08 Lab]$ ls alphafold3 conda jharvard software spack
Pros
- Quick
- Does not require an additional tool/app
Cons
- Learning curve for those not familiar with the command line interface
- No graphical user interface (no point and click, all interactions are done via the keyboard)
Option 2: Filezilla
FileZilla is a free and open-source SFTP (secure file transfer protocol) client. You can find step-by-step instructions on how to use FileZilla in the SFTP file transfer using Filezilla (Mac/Windows/Linux) documentation.
Pros
- Graphical user interface (GUI)
Cons
- When you open a file located on the cluster to edit it, it downloads to your local machine, saves it, and then uploads it back to the cluster — this process can sometimes be slow.
Option 3: Open OnDemand (OOD)
- Connect to the FASRC VPN (see VPN setup)
- Go to the Open OnDemand dashboard
- On the top menu, click “Files” tab
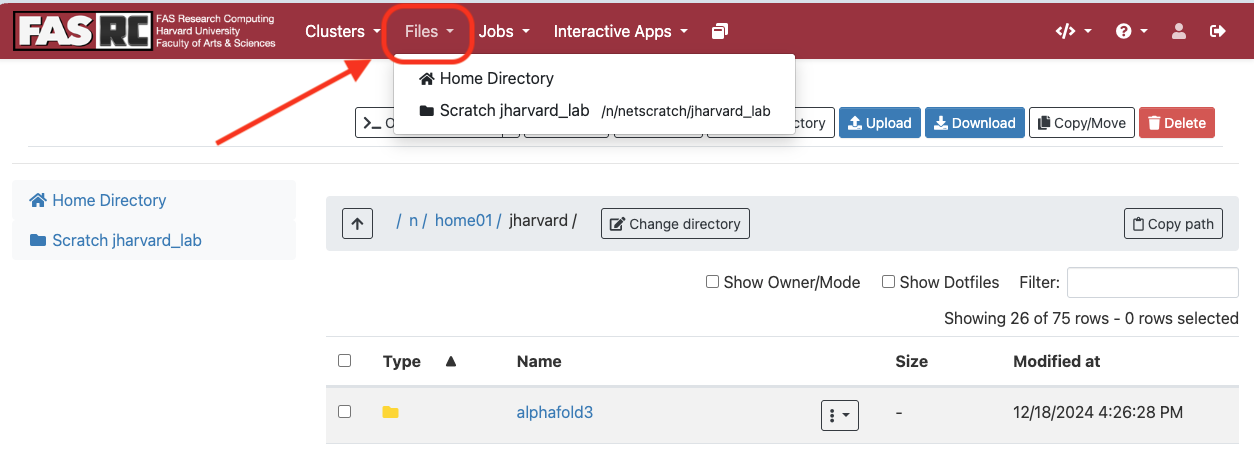
- Double-click on the folder that you would like to open
- To open a file or edit a file, click on the 3 dots, then select “View” or “Edit”

Pros
- Graphical user interface (GUI)
- You may open files and edit them without having to download them to your local machine
Cons
- If you have a lot of files (hundreds of files) in one folder, OOD may become unresponsive. If this happens, use a different method, as OOD is not designed to browse hundreds or thousands of files. Then, create more folders and spread files into the newly created folders
- Requires connection to the FASRC VPN
Option 4: Globus
Refer to the Globus File Transfer documentation.
Pros
- Graphical user interface (GUI)
- Fast
- Can also be used to transfer files to your local computer and other institutions that have Globus
Cons
- May take some time to get familiar with Globus
- Viewing/opening files is not great
- Not recommended if transferring files within the same filesystem (e.g., from a folder in holylabs to another folder in holylabs; in this case, use the command
mv)
How do I transfer files?
Moving data between storage offerings can be a challenge. There are several commands and tools you can use to move data back and forth.
Within the cluster
Option 1: Command-line interface
For options using the command-line interface, refer to Transferring Data on the Cluster documentation (cp, mv, rsync, fpsync commands)
Option 2: Open OnDemand Remote Desktop
This is a good option if you need to transfer files to/from your home directory because Globus does not have access to home directories.
- Go to the Open OnDemand dashboard
- Start a Remote Desktop session (for how to start, see Remote Desktop documentation)
- After the Remote Desktop session starts, open a File Manager by clicking on the “Home” folder on the Desktop. (Alternatively, you can use the top left menu: click “Applications” -> “File Manager”
- Navigate to the source folder (the location where the file is being copied from)
- Open another File Manager window
- Navigate to the destination folder (the location where you would like to paste the file)
- By now, you should have something similar to
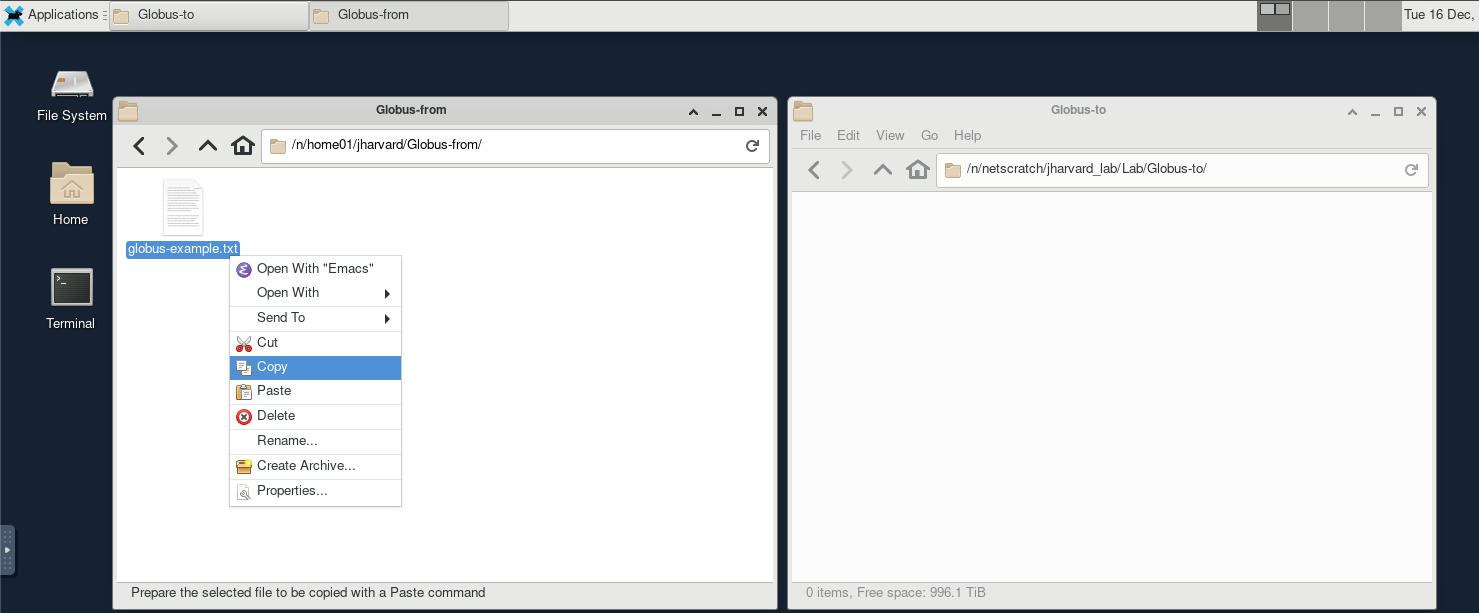
- Left window (source):
/n/home01/jharvard/Globus-from/ - Right window (destination)
/n/netscratch/jharvard_lab/Lab/Globus-to
- Left window (source):
- Right-click the source file -> Copy
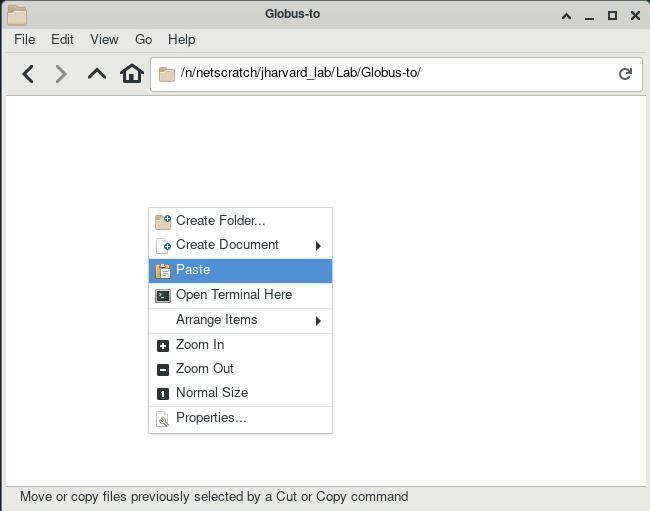
- Right-click the destination window -> Paste
- You may drag and drop files from the source window to the destination window, but this is similar to cut-and-paste.
Option 3: Globus
Only recommended if you are transferring between different file systems (e.g. holylabs to/from netscratch, group storage to/from netscratch). Globus is not recommended for transfers in the same filesystem (e.g. from a folder in holylabs to a different folder in holylabs).
See example 4 in the Globus File Transfer documentation.
Outside of the cluster
To transfer data outside of the cluster, refer to the Transferring Data Externally documentation. There are different ways in which to transfer data to and from research computing facilities. The appropriate choice will depend on the size of your data, your need to secure it, and also who you wish to share it with.
How do I know how much storage is available?
Option 1: Command-line interface
The FASRC quota command provides storage limit and usage information for all FASRC storage options except Cold Storage (Tape). Additional information can be found on the Checking quota and usage page.
Option 2: Coldfront
You may use Colfront to see your lab’s storage allocation and usage (note that Lustre filesystems, Tier 0, may show different amounts). Refer to the ColdFront User Guide documentation.
Help
Who do I contact if I have questions?
Please email rchelp@rc.fas.harvard.edu if you have any questions!
