Search Docs by Keyword
R and RStudio
Description
R is a language and environment for statistical computing and graphics. There are several options to use R on the FASRC clusters.
Of those options, the FASRC recommended method is the RStudio Server stand-alone app on Open OnDemand.
We recommend using RStudio Server on Open OnDemand because it is the simplest way to install R packages (see RStudio Server). We only recommend R module and RStudio Desktop if you:
- plan to run mpi/multi-node jobs
- need to choose specific compilers for R package installation
- are an experienced user, who
- knows how to compile software from source
- has too much time on their hands
- likes to take risks that often don’t payoff ( Hey, there is always RStudio on Open OnDemand, right? )
Should you require it, we offer ( “at your own risk” ):
- R module from command line interface
- R module + RStudio Desktop application on Remote Desktop app on Open onDemand
- R in Jupyter
- R with Spack
Usage
RStudio on Open OnDemand
Use RStudio on Open OnDemand to reduce your stress and that of those around you. Here is a short 5 minute video to get you started:
RStudio Server
RStudio Server is our go-to RStudio app because it contains a wide range of precompiled R packages from bioconductor and rocker/tidyverse. This means that installing R packages in RStudio Server is pretty straightforward. Most times, it will be sufficient to simply:
> install.packages("package_name")
This simplicity was possible because RStudio Server runs inside a Singularity container, meaning that it does not use the host operating system (OS). For more information on Singularity, refer to our Singularity on the cluster docs.
Important notes:
- User-installed R libraries will be installed in
~/R/ifxrstudio/\<IMAGE_TAG\> - This app contains many pre-compiled packages from bioconductor and rocker/tidyverse.
- FAS RC environment modules (e.g.
module load) and Slurm (e.g.sbatch) are not accessible from this app. - For the RStudio with environment module and Slurm support, see RStudio Desktop
This app is useful for most applications, including multi-core jobs. However, it is not suitable for multi-node jobs. For multi-node jobs, the recommended app is RStudio Desktop.
FASSE cluster additional settings
If you are using FASSE Open OnDemand and need to install R packages in RStudio Server, you will likely need to set the proxies as explained in our Proxy Settings documentation. Before installing packages, execute these two commands in RStudio Server:
> Sys.setenv(http_proxy="http://rcproxy.rc.fas.harvard.edu:3128") > Sys.setenv(https_proxy="http://rcproxy.rc.fas.harvard.edu:3128")
Package Seurat
In RStudio Server Release 3.18, the default version for umap-learn is 0.5.5. However, this version contains a bug. To resolve this issue, downgrade to umap-learn version 0.5.4:
> install.packages("Seurat")
> reticulate::py_install(packages = c("umap-learn==0.5.4","numpy<2"))
And test with
> library(Seurat)
> data("pbmc_small")
> pbmc_small <- RunUMAP(object = pbmc_small, dims = 1:5, metric='correlation', umap.method='umap-learn')
UMAP(angular_rp_forest=True, local_connectivity=1, metric='correlation', min_dist=0.3, n_neighbors=30, random_state=RandomState(MT19937) at 0x14F205B9E240, verbose=True)
Wed Jul 3 17:22:55 2024 Construct fuzzy simplicial set
Wed Jul 3 17:22:56 2024 Finding Nearest Neighbors
Wed Jul 3 17:22:58 2024 Finished Nearest Neighbor Search
Wed Jul 3 17:23:00 2024 Construct embedding
Epochs completed: 100%| ██████████ 500/500 [00:00]
Wed Jul 3 17:23:01 2024 Finished embedding
R, CRAN, and RStudio Server pinned versions
FASRC RStudio server pins R, CRAN, and RStudio Server versions to a specific date to ensure R package compatibility. Therefore, we strongly recommend using > install.packages("package_name") with no repos argument specified.
For example, in Release 3.20:
> install.packages("parallelly")
Installing package into ‘/n/home12/jharvard/R/ifxrstudio/RELEASE_3_20’
(as ‘lib’ is unspecified)
trying URL 'https://p3m.dev/cran/__linux__/noble/2025-02-27/src/contrib/parallelly_1.42.0.tar.gz'
Content type 'binary/octet-stream' length 537560 bytes (524 KB)
==================================================
downloaded 524 KB
* installing *binary* package ‘parallelly’ ...
* DONE (parallelly)
The downloaded source packages are in
‘/tmp/Rtmp1AiMaa/downloaded_packages’
Above, the package is downloaded from https://p3m.dev/cran/__linux__/noble/2025-02-27/src/contrib/parallelly_1.42.0.tar.gz Note the date 2025-02-27 and not latest.
For more details see Rocker project which is the base image for FASRC’s RStudio Server.
Advanced Installation: the R package latest version ( not recommended )
This following approach is not recommended, but if you need to build the latest version of a package from source for some reason, you may specify the repo, version, or install from github. Do note that using this approach is very tricky and will likely break R package dependencies. Please do not do this. Kittens will explode.
repos example:
install.packages("rstan", repos = "https://cloud.r-project.org")
install_github example:
> require(remotes)
> install_github("wadpac/GGIR", ref = "3.2-10")
Use R packages from RStudio Server in a batch job
The RStudio Server OOD app hosted on Cannon at rcood.rc.fas.harvard.edu and FASSE at fasseood.rc.fas.harvard.edu runs RStudio Server in a Singularity container (see Singularity on the cluster). The path to the Singularity image on both Cannon and FASSE clusters is the same:
/n/singularity_images/informatics/ifxrstudio/ifxrstudio:RELEASE_<VERSION>.sif
Where <VERSION> corresponds to the Bioconductor version listed in the “R version” dropdown menu. For example:
R 4.2.3 (Bioconductor 3.16, RStudio 2023.03.0)
uses the Singularity image:
/n/singularity_images/informatics/ifxrstudio/ifxrstudio:RELEASE_3_16.sif
As mentioned above, when using the RStudio Server OOD app, user-installed R packages by default go in:
~/R/ifxrstudio/RELEASE_<VERSION>
This is an example of a batch script named runscript.sh that executes R script myscript.R inside the Singularity container RELEASE_3_16:
#!/bin/bash
#SBATCH -c 1 # Number of cores (-c)
#SBATCH -t 0-01:00 # Runtime in D-HH:MM
#SBATCH -p test # Partition to submit to
#SBATCH --mem=1G # Memory pool for all cores (see also --mem-per-cpu)
#SBATCH -o myoutput_%j.out # File to which STDOUT will be written, %j inserts jobid
#SBATCH -e myerrors_%j.err # File to which STDERR will be written, %j inserts jobid
# set R packages and rstudio server singularity image locations
my_packages=${HOME}/R/ifxrstudio/RELEASE_3_16
rstudio_singularity_image="/n/singularity_images/informatics/ifxrstudio/ifxrstudio:RELEASE_3_16.sif"
# run myscript.R using RStudio Server signularity image
singularity exec --cleanenv --env R_LIBS_USER=${my_packages} ${rstudio_singularity_image} Rscript myscript.R
To submit the job, execute the command:
sbatch runscript.sh
R and RStudio on Windows
See our R and RStudio on Windows page.
Advanced Usage: Not Better, Not Faster and Not Recommended *
( * Fine, it could be faster if you really know what you are doing. Still not recommended.)
These options are for users familiar with software installation from the source, where you choose compilers and set your environmental variables. If you are not familiar with these concepts, we highly recommend using RStudio Server instead.
WARNING: If you got really good at using RStudio with Open OnDemand, and think you are expert now– think you are advanced now– You are not. Go back to Open OnDemand where you were well supported and experienced near effortless success.
R module
To use R module, you should first have taken our Introduction to the FASRC training and be familiar with running jobs on the cluster. R modules come with some basic R packages. If you use a module, you will likely have to install most of the R packages that you need.
To use R on the FASRC clusters, load R via our module system. For example, this command will load the latest R version:
module load R
If you need a specific version of R, you can search with the command
module spider R
To load a specific version
module load R/4.2.2-fasrc01
For more information on modules, see the Lmod Modules page.
To use R from the command line, you can use an R shell for interactive work. For batch jobs, you can use R CMD BATCH and RScript commands. Note that these commands have different behaviors:
R CMD BATCH- output will be directed to a
.Routfile unless you specify otherwise - prints out input statements
- cannot output to STDOUT
- output will be directed to a
RScript- output and errors are directed to to STDOUT and STDERR, respectively, as many other programs
- does not print input statements
For slurm batch examples, refer to FASRC User_Codes Github repository:
Examples and details of how to run R from the command line can be found at:
- Stack Overflow post
- R doc pages
- O’Reilly Books Online for Harvard (valid Harvard ID required) and then search for “R programming” or “R cookbook”
R Module + RStudio Desktop
RStudio Desktop depends on an R module. Although it has some precompiled R packages that comes with the R module, it is a much more limited list than the RStudio Server app.
RStudio Desktop runs on the host operating system (OS), the same environment as when you ssh to Cannon or FASSE.
This app is particularly useful to run multi-node/mpi applications because the you can specify the exact modules, compilers, and packages that you need to load.
See how to launch RStudio Desktop documentaiton.
R in Jupyter
To use R in Jupyter, you will need to create a conda/mamba virtual environment and install packages jupyter and rpy2 , which will allow you to use R in Jupyter.
Step 1: Request an interactive job
salloc --partition test --time 02:00:00 --ntasks=1 --mem 10000
Step 2: Load python module, set environmental variables, and create an environment with the necessary packages:
module load python/3.10.13-fasrc01 export PYTHONNOUSERSITE=yes mamba create -n rpy2_env jupyter numpy matplotlib pandas scikit-learn scipy rpy2 r-ggplot2 -c conda-forge -y
See Python instructions for more details on Python and mamba/conda environments.
After creating the mamba/conda environment, you will need to load that environment by selecting the corresponding kernel on the Jupyter Notebook to start using R in the notebook.
Step 3: Launch the Jupyter app on the OpenOnDemand VDI portal using these instructions.
You may need to load certain modules for package installations. For example, R package lme requires cmake. You can load cmake by adding the module name in the field “Extra Modules”:
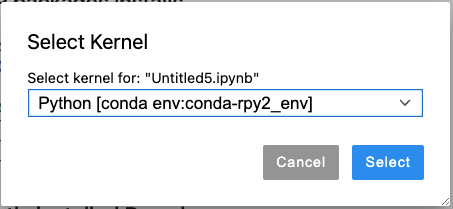
Step 4: Open your Jupyter notebook. On the top right corner, click on “Python 3” (typically, it has “Python 3”, but it may be different on your Notebook). Select the created conda environment “Python [conda env:conda-rpy2_env]”:
Alternatively, you can use the top menu: Kernel -> Change Kernel -> Python [conda env:conda-rpy2_env]
Step 5: Install R packages using a Jupyter Notebooks
Refer to the example Jupyter Notebook on FASRC User_Codes Github.
R with Spack
Step 1: Install Spack by following our Spack Install and Setup instructions.
Step 2: Install the R packages with Spack from the command line. For all R package installations with Spack, ensure you are in a compute node by requesting an interactive job (if you are already in a interactive job, there is no need to request another interactive job):
[jharvard@holylogin spack]$ salloc --partition test --time 4:00:00 --mem 16G -c 8
[jharvard@holy2c02302 spack]$ spack install package_name # install software
[jharvard@holy2c02302 spack]$ spack load package_name # load software to your environment
[jharvard@holy2c02302 spack]$ R # launch R
> library(package_name) # load package within R
R Parallel
This is covered at R Parallel here on User Docs, and EXTENSIVELY in User Docs/Parallel_Computing/R.
Troubleshooting
Files that may configure R package installations
~/.Rprofile~/.Renviron~/.bashrc~/.bash_profile~/.profile~/.config/rstudio/rstudio-prefs.json~/.R/Makevars